|
||
|
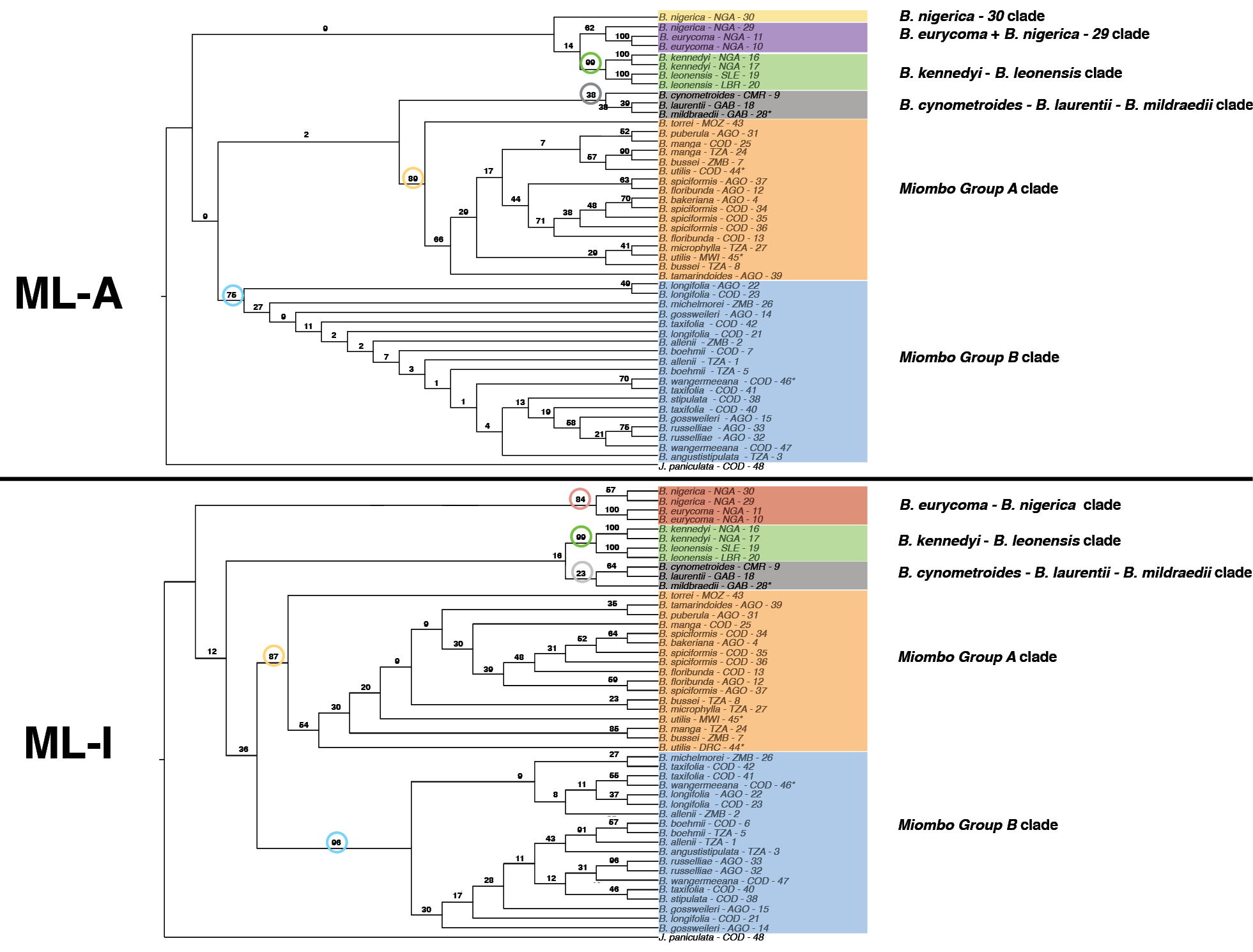
Maximum Likelihood phylogenetic inferences of Brachystegia species using rDNA sequences and two different coding schemes. Cladograms were produced using RAxML-NG software (Kozlov et al. 2019) and intra-individual site polymorphisms (2ISPs) were coded following the IUPAC nomenclature. 2ISPs are either considered as ambiguous (ML-A) or are coded as state into the substitution model (ML-I). Bootstrap supports (BS) are indicated on each branch. |