|
||
|
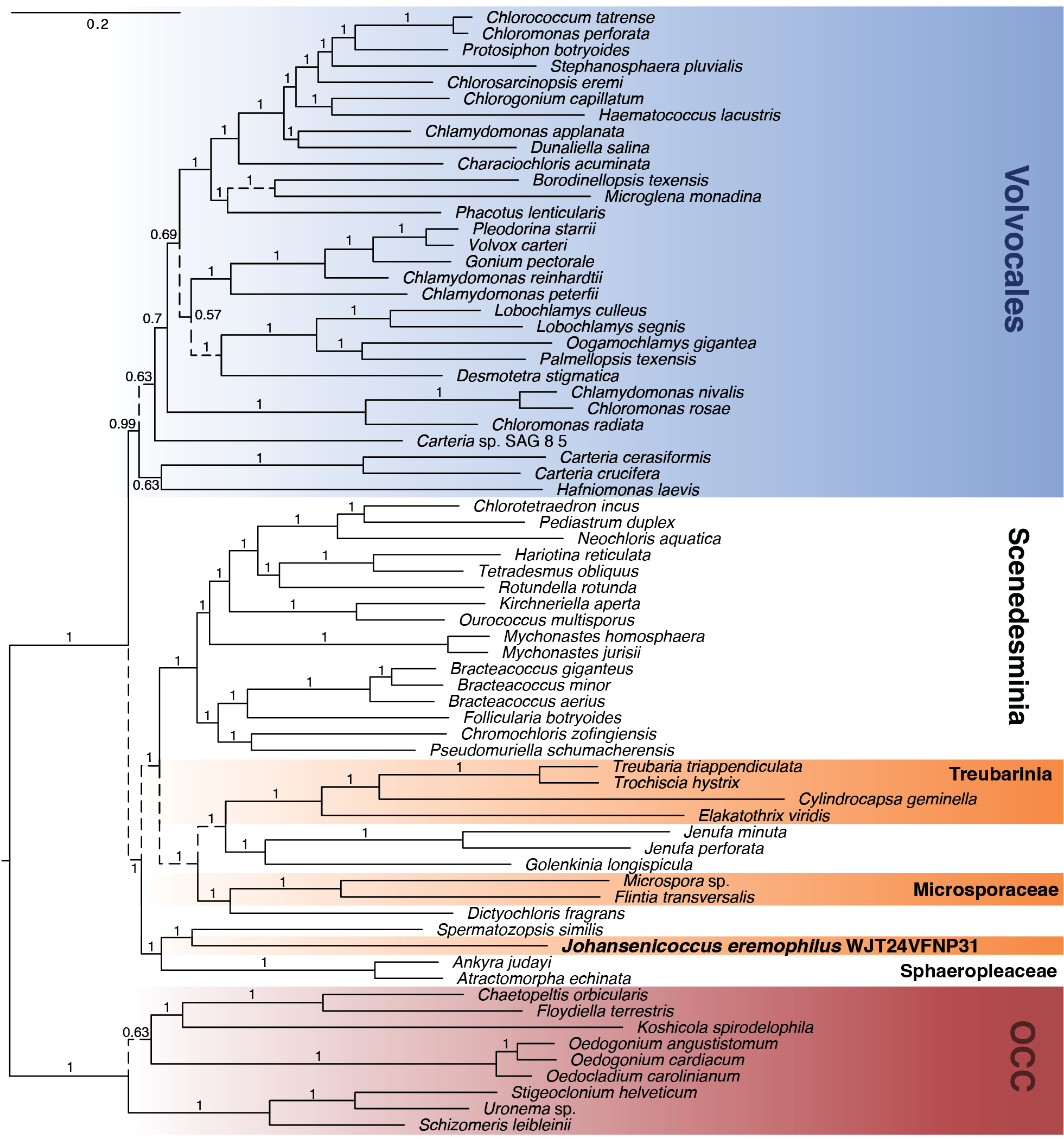
Bayesian consensus phylogeny based on an analysis of 59 concatenated protein-coding chloroplast genes, with the nucleotide data partitioned by codon position. Numbers on branches represent Bayesian Posterior Probability. Solid branches represent relationships that were also recovered in the corresponding Bayesian analysis of the amino acid data. Dashed branches indicate relationships that were not recovered in the amino acid analysis. Scale bar represents the estimated number of nucleotide substitutions per site. |